Readme of GalaXimView¶
GalaXimView (for Galaxies Simulations Viewer) is designed to visualise simulations which use particles, providing notably a rotatable 3D view and corresponding projections in 2D, together with a way of navigating through snapshots of a simulation keeping the same projection.
GalaXimView was written by Anaelle Halle.
Questions and feedback are welcome.
Requirements¶
GalaXimView requires python 3 (likely >=3.6) and the following python libraries:
matplotlib >= 3.2.0
numpy >= 1.17.5
scipy >= 1.3.2
h5py >= 2.6.0 (only for support of HDF5 input reading)
You can check which versions you have by importing modules (e.g. import matplotlib) and executing
modulename.__version__ (e.g. matplotlib.__version__) in a python console.
Personal way of installing or updating libraries is, for example for h5py: pip3 install h5py --upgrade
Installation¶
Cloning the git repository by:
git clone https://gitlab.obspm.fr/ahalle/galaximview.git
will create the ‘galaximview’ directory at the cloning location. The ssh cloning appears to be problematic (but https cloning with the command line above or by clicking on the gitlab option works fine). You can also download an archive on the gitlab website.
You should add this galaximview directory to your python path. For example, in your .bashrc or in your .bash_profile (or .profile):
export PYTHONPATH="/path/of/directory:${PYTHONPATH}"
where ‘/path/of/directory’ should be replaced with the proper path of the galaximview directory (the .bashrc should then be
sourced by executing source .bashrc or, if you have edited the .bash_profile or .profile, you should log out
and in again).
Or, in a python console:
import sys
sys.path.extend(['/path/of/directory'])
Then typing in a python console:
import galaximview
should not raise any error.
Running¶
GalaXimView needs to be run interactively, in an interactive ipython console (especially to use matplotlib buttons), opened with
ipython(if you do not already have ipython installed, you can install it following https://ipython.org/install.html). Note that methods of some classes such as the Snap class can be used separately to perform computations or plot simple figures.
GalaximView has been tested to work with the following backends: ‘GTK3Agg’, ‘Qt4Agg’, ‘Qt5Agg’, ‘TkAgg’. You can check which backend you are using by:
import matplotlib matplotlib.get_backend()
and change it if desired by exiting the ipython console and in a new one, typing, for example:
import matplotlib matplotlib.use('Qt5Agg')
Personal preference is for ‘Qt5Agg’ which may be installed by
pip3 install pyqt5 --upgrade
(In case of graphical problem, you can try another backend.)
In the script launchviewer.py, ‘example_path_to_simu_directory’ should be set to a path to the directory containing the snapshots of the simulation you want to visualise. The viewer can then be launched by typing, in the ipython console:
run /path/to/launchviewer.py
with of course ‘/path/to’ replaced to the proper path, or removed if the script is run from inside its directory. Please read the ‘Examples’ section below to run the script with example simulations.
GalaXimView runs more quickly on simulations with a moderate number of particles (up to a few millions). So far, it does not include any parallelization and could thus crash if the computer on which it is run does not have enough RAM. As it heavily uses a graphics backend, it may be easier to run it on a local computer than on a server. Some mounting of external partitions such as done by sshfs can be used (if it fits to the server users guidelines).
Examples¶
You can download some small simulations taken from the examples of Gadget-2 / Gadget-4 (Springel V., Pakmor R., Zier O., Reinecke M., MNRAS):
(a subset of snapshots of) a simulation with gas and star formation (173 Mb):
wget https://vm-weblerma.obspm.fr/~ahalle/data/galaximview/CollidingGalsSFR-G4-hdf5.tar.gz
a simulation with only stars and dark matter (12 Mb):
wget https://vm-weblerma.obspm.fr/~ahalle/data/galaximview/G2-galaxy-G4run-G1G2format.tar.gz
The script ‘launchviewer.py’ visualises these simulations assuming that they are stored (extracted from the archives) in the ‘example_simulations’ directory (with a path ‘../example_simulations’ relative to the current directory).
Once you have downloaded and extracted the files, to run GalaximView on one of these simulations you should thus:
edit launchviewer.py to set which simulation you want to visualise and to set the path to the directory containing the snapshots of the simulation (absolute path or path relative to the location from which you open
ipython).open
ipythonand executerun /path/to/launchviewer.py
Expanding¶
The code can be adapted to other input types and plotting of user-customised quantities can be added by creating new buttons.
To add another quantity in the snap method, you can:
If required, add reading of snapshot field in the proper reading routine, choose a string to refer to the field and make it a member of the data dictionary returned by the routine.
Choose a string referring to the quantity to be added and add a case in the if loop of Snap.get_array_from_string(). Add the string to Snap.listquantities (optional).
If proper units conversion is desired, edit BaseUnitsDefs.get_conversion_factor_from_string(), BaseUnitsDefs.get_dic_of_labels() and BaseUnitsDefs.get_dic_of_labels_units(). If not, the code will run but with warnings.
Then, to see this quantity in 2D plots (for example), you can:
Add a case in the if loop of GalViewer.plot_2d().
Add a string to refer to the new button and its location for example in dic_new_buttons of GalViewer.create_buttons, then a label to be shown on the button and an action to be done when clicking on it in ButtonsActions (editing, for example, the ButtonsActions.action_nothing() method added as an example of a ‘Nothing’ button.)
Documentation¶
Documentation of the different parts of the code can be found on the website of the code (or by clicking on ‘galaximview package’ on the left menu if you are viewing this README from the sphinx generated html documentation.)
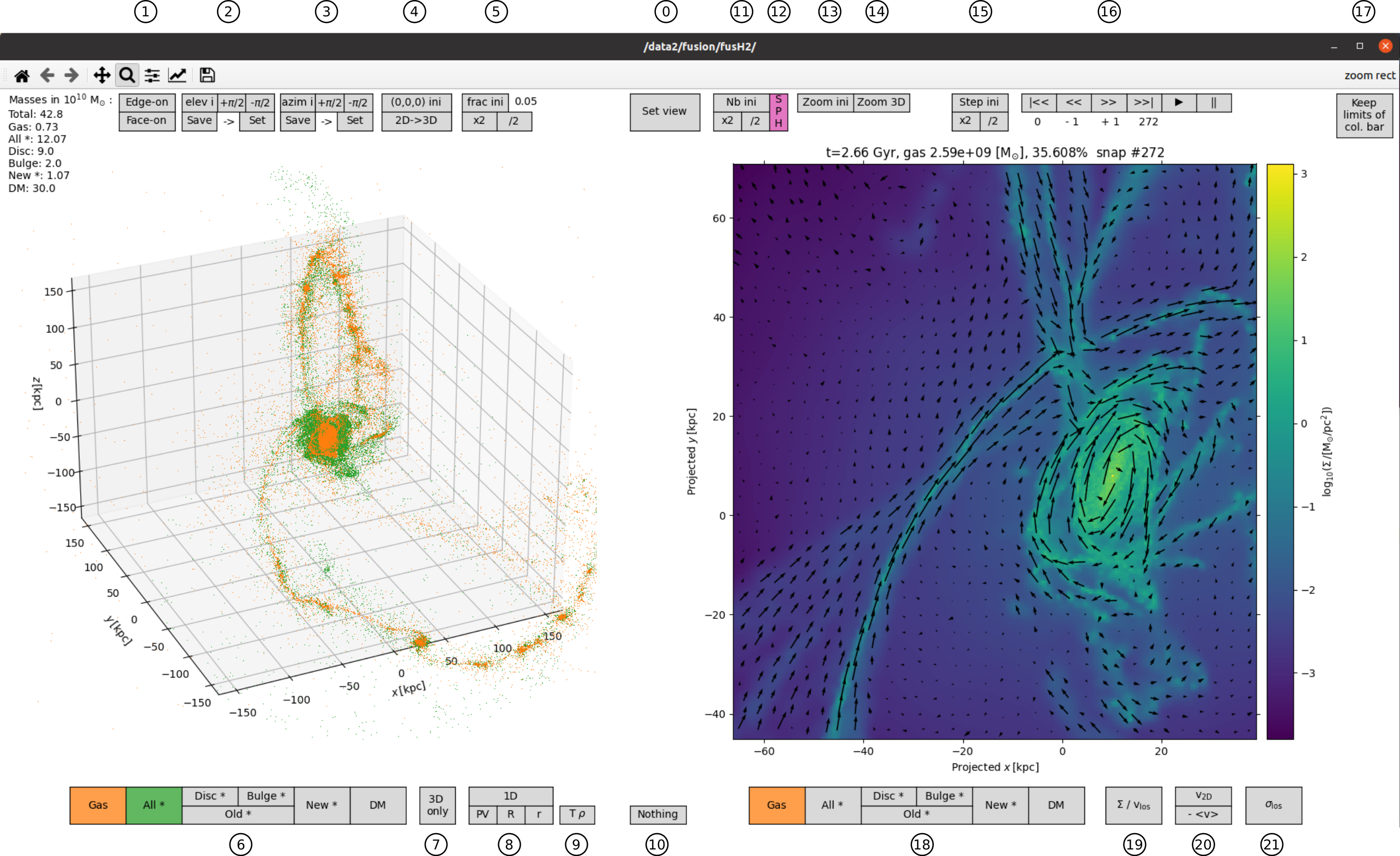
The 3D pot is rotatable and zoomable with the normal graphical backend buttons. The 2D plot is also zoomable with the normal graphical backend buttons and can be replotted by clicking on Set view.
0: Set view: To set the 2D projection corresponding to 3D plot and to replot the 2D plot after zooming in or out (with normal graphical backend buttons)
1: Edge-on: To get a line-of-sight in the (x,y) plane. Face-on: To get a line-of-sight orthogonal to the (x,y) plane
2: elev i: To get the initial elevation. +pi/2: To increase the elevation of pi/2. +-pi/2: To decrease the elevation of pi/2. Save: To temporarily save the elevation. Set: to reset the elevation to the saved value previously obtained by Save (this can be used if one wants to change only the azimuth freely by rotating the plot but while recovering the exact same elevation).
3: azim i: To get the initial azimuth. +pi/2: To increase the azimuth of pi/2. +-pi/2: To decrease the azimuth of pi/2. Save: To temporarily save the azimuth. Set: to reset the azimuth to the saved value previously obtained by Save (this can be used if one wants to change only the elevation freely by rotating the plot but while recovering the exact same azimuth).
4: (0,0,0) ini: To reset the origin of the 3D plot to its initial value. 2D->3D To set the origin from the 2D plot (useful to easily zoom in on particles far from the origin (0,0,0))
5: frac ini: To reset the fraction of shown particles in the 3D plot t its original value (5% by default). x2: To multiply the fraction of shown particles by 2. /2: To divide the fraction of shown particles by 2.
6: Gas, All *, Disc *, Bulge *, Old *, New *, DM: To show or remove particle of the corresponding component on the 3D plot.
7: 3D only: To have only the 3D plot on the left part of the window (by default).
8: 1D: To get mass, velocity and velocity dispersion histograms in 1D. PV: as a function of x. R: in cylindrical geometry. r: in spherical symmetry.
9: T rho: To get the 2D distribution of temperature vs density (and marginal distributions) of the gas.
10: Nothing: Does nothing, just shown as example of potential new button in the code.
11: Nb ini: To reset the number of bins (of equal size along the x and the y axes) to the initial value (default is 256 on the x axis). x2: To multiply the number of bins of the x axis by 2. /2: To divide the number of bins of the x axis by 2.
12: SPH: (slow) To render density, line-of-sight velocity and line-of-sight velocity dispersion for gas using the smoothing lengths of the gas particles. The algorithm is rather slow for the moment.
13: Zoom ini: To reset the 2D box x and y limits to their original values.
14: Zoom 3D: To set the size of the 2D box so that it corresponds to what is seen on the 3d plot.
15: Step ini: To set the step (between snapshots) to the initial value (default = 1). x2: To multiply the step by 2. /2: To divide the step by 2.
16: Player-like time control buttons: |<< To go to the first snapshot. >>| To go to the last snapshot. << To go back by one step. >> To go forward by one step. |> To ‘play’ the snapshots forward. || To pause (stop at the current snapshot) the play.
17: Keep the limits of col. bar: keep the limits of the colour bar when replotting in 2D.
18: Gas, All *, Disc *, Bulge *, Old *, New *, DM: To show the corresponding component on the 2D plot.
19: Sigma/vlos: To show either the surface density or line-of-sight velocity.
20: v2D: To show a quiver plot (arrows) of the velocity on the plane of the 2D projection. -<v>: To remove the average value of x and y velocities in the 2D box when plotting the quiver plot (useful for example To show the velocity field in a part of a snapshot)
21: sigma los: To show the dispersion of the line-of-sight velocity.
Note that if the viewer is stored as gv as in the launchviewer.py example file, the 2d and 3d plots can also be accessed and modified in the console by editing gv.ax2d and gv.ax3d.